1. Introduction
CNIT (Coding-NonCoding Identifying Tool) software is a powerful signature tool to effectively distinguish between protein-coding and non-coding sequences by profiling adjoining nucleotide triplets ANT based on sequence intrinsic composition, especially for classification of incomplete transcripts and sense-antisense transcript pairs. Last version of CNCI1 is widely used by worldwide researchers. For better serve for scientific community and to make users distinguish transcripts more conveniently, we update CNIT to CNIT. It can discriminate the coding and non-coding transcripts faster, more accurately, in more species, especially for plants. Here, we provide an online version of CNIT.
2. CNIT Online Input
CNIT accepts RNA transcript sequences in fasta format.
2.1 Submit requirement
Fasta format:
Size requirement: Less than 10000 lines in input box and no line limitation in batch model. Maximum allowable upload file size is 50 Mb.
Name requirement: Sequence names beginning with ‘>’ symbol are required.
Sequence requirement: Only characters in DNA and RNA sequences are case ignoring, such as ATCGUatcgu.
GTF format are supported when you install the CNIT standalone version on most Linux-based operating systems not in Web temporarily.
2.2 How to submit the RNA transcript sequences?
There are two ways to submit RNA transcript sequences:
1) Paste RNA sequences in fasta format into the big input box at the home page.
2) Upload fasta file by the batch operation.
3. CNIT Results Output
The results will be stored on our server for seven days, you can retrieve your result via the job-ID link.
3.1 CNIT results: html view
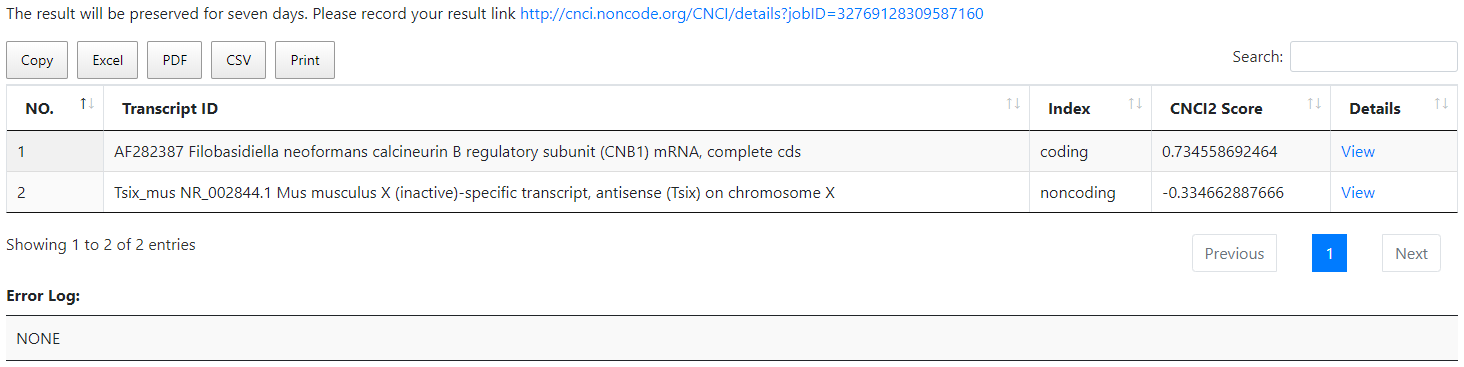
CNIT results html view gives an overview of coding status of the input sequences. Each row corresponds to one input sequence. The columns show the Transcript ID, the coding/noncoding classification label (Index), the coding probability score (CNIT Score) The results can be copied, printed and downloaded directly from our server in different file formats such Excel, PDF, csv.
3.2 CNIT results details

Results details consist of the most-like CDS (MLCDS) region detail (Figure A), Sequence (Figure B) and CNIT Score Detail Plot: Red line represents the correct transcriptional reading frame and other five lines (blue or green) represent other five reading frames, green line indicates the distribution of the coverage (the right y-axis) of the MLCDS region for each protein-coding transcript across the normalized length, here, we show identification result of coding and noncoding sequence sample (Figure C). Moreover, you blast your sequence in NONCODE database in this page directly.
4. FAQ
The framework of CNIT

The top panel shows the process of a sequence in a testing set. For a given sequence, six MLCDS regions (represented by six lines) are identified from six reading frames (represented by six color arrow lines) using a sliding window and dynamic programming algorithm. Then, an MLCDS region with a maximal S-score is selected to incorporate into an Xgboost. The bottom panel shows the training and classification process. Reliable protein-coding and non-coding sequences are used as a training set, and four features including 67 values are extracted to train Xgboost, which classifies the incorporating sequence into protein-coding or non-coding sequence.
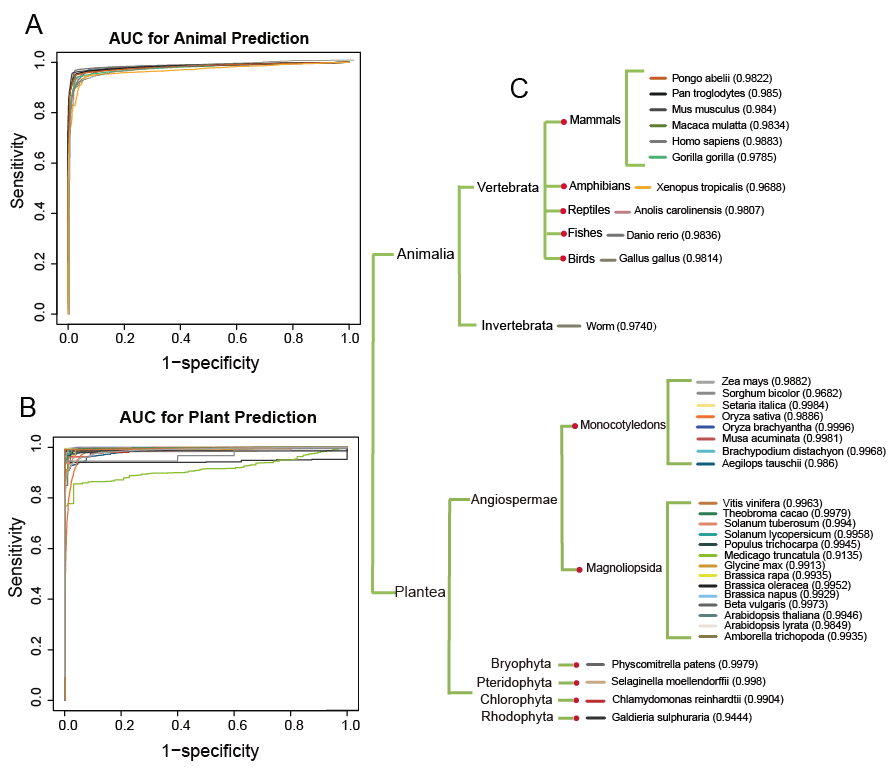
The global prediction for CNIT
Here, we showed the prediction of CNIT for 37 species (11 animal species, 26 plant species) with the corresponding AUC value.
What's new in CNIT?
In comparison with CNCI, CNIT runs∼200 times faster than CNIT and exhibits more accurate in more species, especially for plants, when using Human and Arabidopsis data as training sets. Because CNIT can classify protein-coding and non-coding RNAs solely based on sequence intrinsic composition as CNCI, it is potentially applicable to a variety of species without whole-genome sequence or with poorly annotated information. The last but not the least, the updated model in CNIT can identify the most species in existing software of distinguish transcripts coding probability.
